Archives
- 2026-03
- 2026-02
- 2026-01
- 2025-12
- 2025-11
- 2025-10
- 2023-07
- 2023-06
- 2023-05
- 2023-04
- 2023-03
- 2023-02
- 2023-01
- 2022-12
- 2022-11
- 2022-10
- 2022-09
- 2022-08
- 2022-07
- 2022-06
- 2022-05
- 2022-04
- 2022-03
- 2022-02
- 2022-01
- 2021-12
- 2021-11
- 2021-10
- 2021-09
- 2021-08
- 2021-07
- 2021-06
- 2021-05
- 2021-04
- 2021-03
- 2021-02
- 2021-01
- 2020-12
- 2020-11
- 2020-10
- 2020-09
- 2020-08
- 2020-07
- 2020-06
- 2020-05
- 2020-04
- 2020-03
- 2020-02
- 2020-01
- 2019-12
- 2019-11
- 2019-10
- 2019-09
- 2019-08
- 2019-07
- 2019-06
- 2019-05
- 2019-04
- 2018-07
-
br Material and methods br Results
2021-03-02
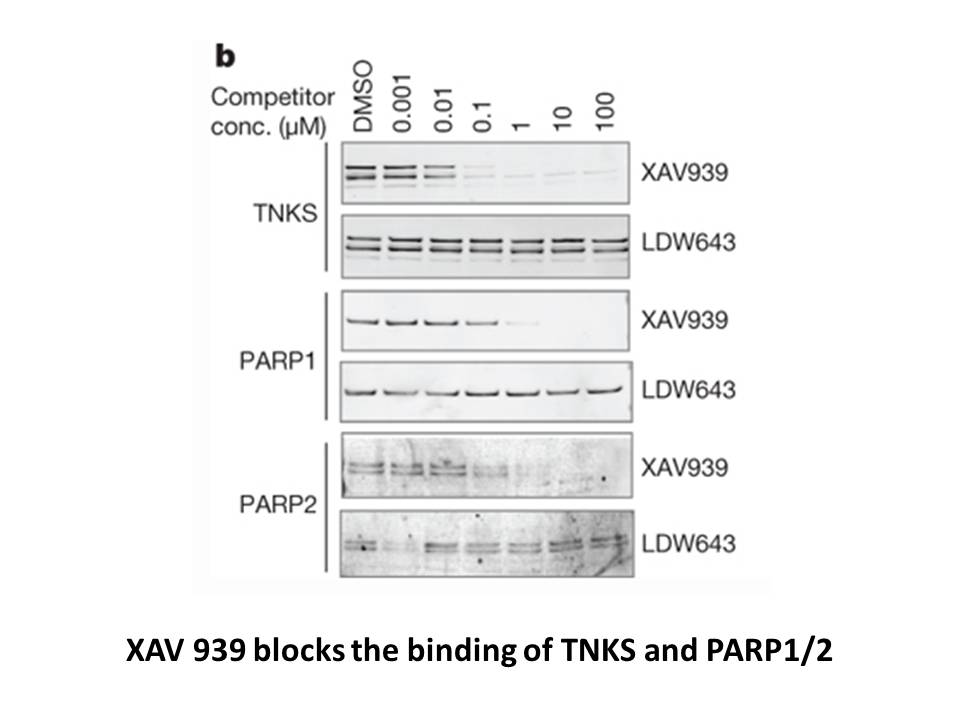
Material and methods Results and discussion Conclusions Prostanoid-E receptor selective antagonists that inhibit EP2 or EP4 receptor activities may be used as a pharmacological strategy to limit cyst formation and ADPKD progression. Our study follows on from our previous observations of the
-
It was hypothesized that if modafinil acts primarily
2021-03-01
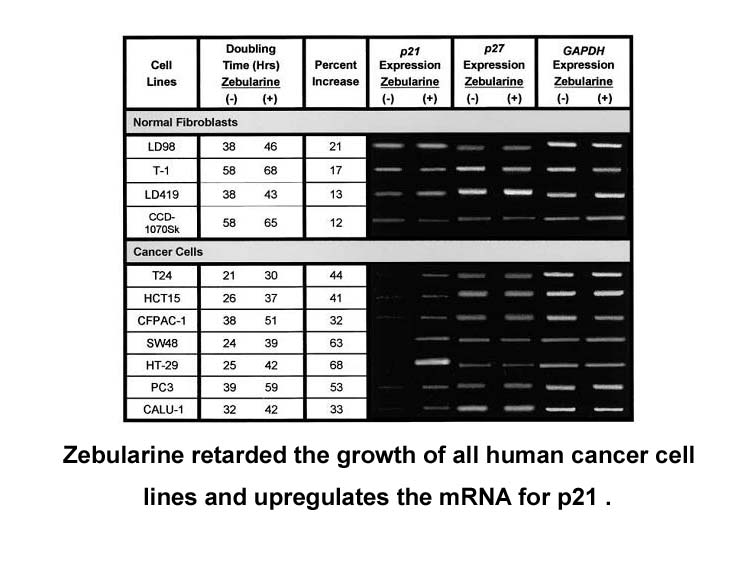
It was hypothesized that if modafinil acts primarily via a noradrenergic mechanism, Dbh −/− mice should be non-responsive since they completely lack NE. In contrast, if modafinil acts mainly through DA systems, these mice should be hypersensitive. Modafinil was tested in Dbh −/− mice using both loco
-
br Acknowledgements We thank Dr James Ritchie and his
2021-03-01
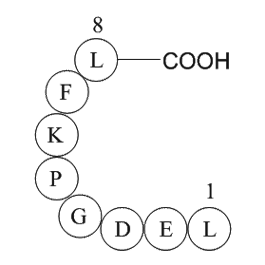
Acknowledgements We thank Dr. James Ritchie and his dedicated staff (Deptartment of Pathology, Emory University) for performing measures of serum VPA levels; Sumitomo Pharmaceuticals (Osaka, Japan) for the generous donation of DOPS, which is required for breeding Dbh −/− mice; Pfizer (Sandwich, K
-
DNA damage represents a persistent threat
2021-03-01
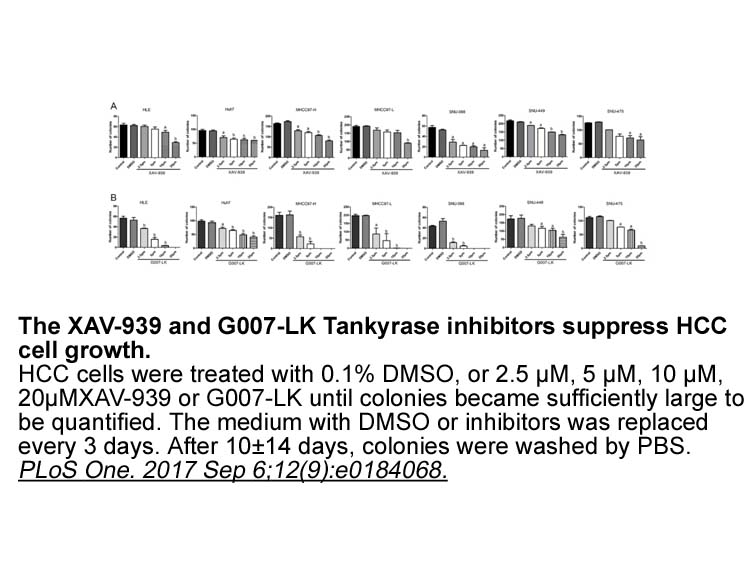
DNA damage represents a persistent threat to genomic stability. A critical link exists between DNA mutation, chromosomal rearrangement and cancer development. In myeloid malignancies, various chromosomal translocations and/or mutations increased cellular reactive oxygen species (ROS), followed by DS
-
The recombinant Scl collagen system has shown capability as
2021-03-01
The recombinant Scl2 collagen system has shown capability as a biomaterial as well because of its adaptability and scalability. Scl2 was functionalized to crosslink into a hydrogel without disrupting its triple helix [130]. The Scl2–hydrogel crosslinking also did not disrupt cell adhesion and integr
-
br The role of chemokine
2021-03-01
The role of chemokine and AD Chemokines are small cytokine proteins that are involved in various immunological and physiological functions. These proteins can be involved in cell migration and localization during homeostasis and inflammation, and as such, they were named chemokines [80]. Chemokin
-
alln Proteins that associate to PCNA are characterized by
2021-03-01
Proteins that associate to PCNA are characterized by having conserved motifs as the PCNA-interacting protein (PIP) motif [29,30], its variant PIP-like motif [31,32] or the APIM motif (Alk B homologue 2 PCNA interacting motif [33,34]); for instance, p21 contains a PIP motif [30], and DNA polymerases
-
Introduction Globally lung cancer has
2021-03-01
Introduction Globally, lung cancer has the highest morbidity and mortality rates of all types of cancer, with 1.5 million patients newly diagnosed annually [1]. Approximately 85% of lung cancer cases are non-small cell lung cancer (NSCLC) [2]. The first-line treatment for NSCLC is surgical resectio
-
We performed three baseline scans in two baboons
2021-03-01
We performed three baseline scans in two baboons and one blocking study by administering 1 mg/kg (i.v) meloxicam 30 min prior to [C]TMI injection (injected activity 175.75 ± 18.5 MBq, mass of unlabeled TMI guggulsterone barrier (BBB) and retained in brain with a somewhat heterogeneous pattern (). T
-
Next to organophosphates carbamates and metals cholinesteras
2021-03-01

Next to organophosphates, carbamates and metals, cholinesterase activity can also be affected by other pesticides, such as newer generation neonicotinoid insecticides (NN). NNs do not inhibit AChE activity via the organophosphate-sensitive binding site of the enzyme, but rather, by acting as a struc
-
br Conclusion and future perspectives LDL cholesterol loweri
2021-03-01
Conclusion and future perspectives LDL cholesterol-lowering therapy is essential for cardiovascular risk reduction. On the background of a high inter-personal variation of cholesterol synthesis and cholesterol lorlatinib australia the efficacy of cholesterol lowering, both, with statins and with
-
The ubiquitin like modifier NEDD
2021-02-27
The ubiquitin-like modifier NEDD8 (NEURAL PRECURSOR CELL EXPRESSED, DEVELOPMENTALLY DOWN-REGULATED 8) uses similar enzymatic machineries for NEDD8 conjugation as ubiquitin (Figure 1) [11, 12, 13, 14]. In plants as well as in other eukaryotes, the number of E3 ligases for NEDD8 and the number of know
-
A previous report demonstrated that DA dependent oxidative s
2021-02-27
A previous report demonstrated that DA-dependent oxidative stress may be the initial event in MA neurotoxicity (Cubells et al., 1994). Oxidative burdens at early stage might be a prerequisite for neurotoxic scenarios induced by 3-FMA, and they are in line with MA case (Shin et al., 2012, Shin et al.
-
While apparent that His Artemis
2021-02-27
While apparent that [His]6-Artemis fractionated over a HAP column is devoid of 5′–3′ exonuclease activity but still retains DNA-PK dependent hairpin-opening activity, we sought to further assess DNA-PK dependent Artemis overhang cleavage activity to ensure all the in vivo, intrinsic enzymatic activi
-
In this work we directly compared the
2021-02-27

In this work, we directly compared the effects on transcription by E. coli RNAP of several types of lesions commonly found in the genomic DNA: thymine dimer (CPD, cyclobutane pyrimidine dimer); 1,N6-ethenoadenine (εA); abasic site (AP); 8-oxoguanine (8oxoG), and thymine glycol (TG). We for the first
11201 records 396/747 page Previous Next First page 上5页 396397398399400 下5页 Last page